22 September 2024
A research team at the LKS Faculty of Medicine, the University of Hong Kong (HKUMed), has achieved a significant breakthrough in understanding how coronaviruses replicate. The pioneering study revealed the role of a specialised structure known as the double-membrane vesicle (DMV) pore in facilitating the movement of newly synthesised viral RNA into host cells. The researchers resolved the molecular architecture of the SARS-CoV-2 pore complex responsible for viral RNA translocation, which is a crucial step towards designing novel antiviral strategies to inhibit SARS-CoV-2 and other coronaviruses. The groundbreaking findings were recently published in the internationally prestigious scientific journal Nature [link to the publication].
Background
Over the past two decades, severe coronavirus outbreaks in humans, such as SARS, MERS and COVID-19, have significantly impacted global health and economies, highlighting the need for deeper molecular-level research on the coronavirus life cycles, particularly replication mechanisms. Coronaviruses, classified as positive-sense RNA viruses, remodel host cell membranes to create structures like double-membrane vesicles (DMVs), which are essential for RNA synthesis and replication. DMVs concentrate replication factors and shield viral RNA from host defences.
Recently, researchers have identified a pore complex on DMVs in cells infected with coronaviruses like SARS-CoV-2, which may aid in transferring newly synthesised RNA for viral assembly. However, the formation and role of DMV pore complex in supporting replication remain poorly understood. More research is needed to clarify these processes and develop strategies against coronavirus infections.
Research methods and findings
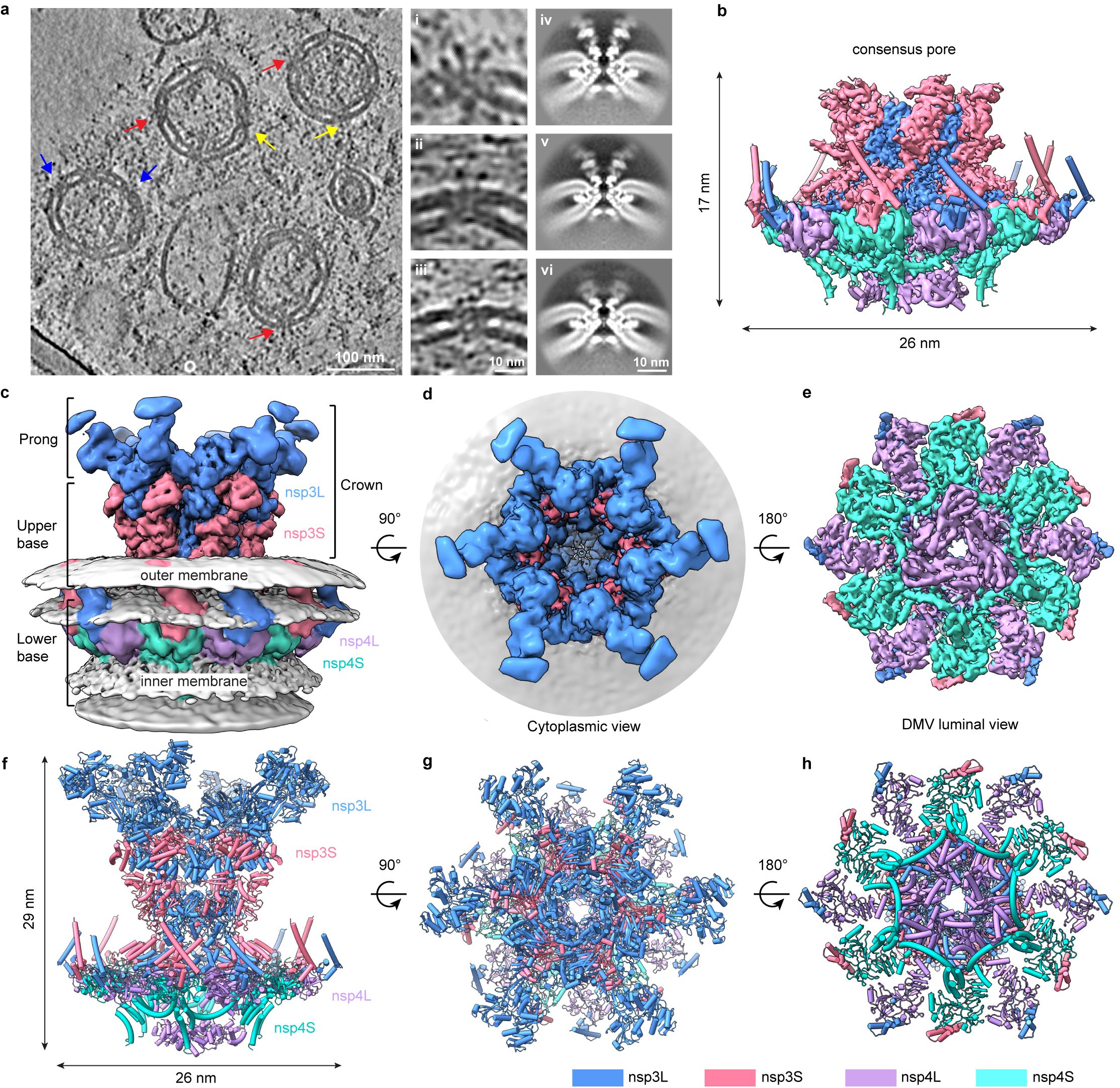
A research team led by Professor Ni Tao, from the School of Biomedical Sciences, and Professor Yuan Shuofeng, from the Department of Microbiology, under the School of Clinical Medicine, both at HKUMed, combined modern structural biology and molecular virology methods to reveal the architecture and function of this viral RNA translocation machinery in the past two years. The team generated and purified DMVs from human embryonic kidney 293F cells and employed cryo-electron tomography (cryo-ET), along with subtomogram averaging, a high-resolution imaging and data-processing technique, to unravel the molecular architecture of the SARS-CoV-2 DMV pore complex formed by non-structural protein nsp3 and nsp4.
The study uncovered the composition and arrangement of the DMV pore complex, which consists of 12 copies each of nsp3 and nsp4 molecular, organised into four concentric stacking hexamer rings, mimicking a miniature nuclear pore complex. A central ring made up of positively charged arginine residues facilitates the viral RNA translocation, which is essential for virus replication. These structures of nsp3-4 pore complex resolve a longstanding question about the quantity and arrangement of these proteins within the DMV pore, uncovering the pathway of membrane zippering and DMV pore formation in the replication organelle. This important discovery sets a foundation for identifying the structure and function of the complete viral replication process.
Significance of the research
The DMV pore complex is currently the only confirmed channel for transporting viral RNA on DMV, and it is essential for viral replication. It is also the only high-resolution DMV pore structure that has been resolved so far. Disrupting the formation of the DMV pore and its ability to transport viral RNA represents a novel approach for inhibiting viral activity. The study will facilitate the rational design of new antiviral strategies against SARS-CoV-2 and other coronaviruses by focusing on blocking DMV pore formation and function, thereby expanding the range of potential therapeutic targets.
Professor Ni Tao expressed his excitement about the findings, stating, ‘Since the SARS outbreak in 2003, research on the coronavirus replication organelle (DMV) and viral RNA translocation mechanism has been a key focus in the field. Over the past 20 years, significant progress has been made, as many individual components of the DMV pore complex have been investigated. However, how these components assemble to become a functional viral RNA translocation pore has remained unknown. With the aid of the state-of-the-art electron microscopy techniques in our newly established Li Ka Shing Cryo-EM Laboratory at HKUMed, we successfully resolved the high-resolution molecular architecture of the coronavirus DMV pore complex.’
The experimental approach developed by Professor Ni and his team highlights the capacity of cryo-ET and subtomogram averaging in studying in vitro systems that recapitulated the in situ environment. The isolated DMVs of the coronavirus provided a valuable in vitro system for investigating the virus-replication mechanism at near-atomic resolution. This research established a foundation for a comprehensive understanding of the complex processes involved in viral replication and transcription, which include the DMV pore complex and RNA replication and transcription machinery.
About the research team
The research team was led by Professor Ni Tao, Assistant Professor, School of Biomedical Sciences at HKUMed, in collaboration with Professor Yuan Shuofeng, Assistant Professor, Department of Microbiology, School of Clinical Medicine, both at HKUMed. Other team members from HKUMed were Huang Yixin, School of Biomedical Sciences; Dr Wang Tongyun, Department of Microbiology, School of Clinical Medicine; Zhong Lijie, Dr Zhang Wenxin and Zhang Yu, School of Biomedical Sciences; and Professor Yu Xiulian, The Hong Kong Polytechnic University. For more information, please visit https://www.tni-lab.org/.
Acknowledgements
The cryo-electron tomography data collection was accessed through the Li Ka Shing Cryo-EM Laboratory under the Centre for PanorOmic Sciences (CPOS) at HKUMed. The study was supported by the Early Career Scheme by the Research Grants Council (RGC); the Guangdong Natural Science Fund; the National Natural Science Foundation of China (NSFC); the NSFC/ RGC Joint Research Scheme; and the Collaborative Research Fund by the RGC.
About the Li Ka Shing Cryo-EM Laboratory
The Li Ka Shing Cryo-EM Laboratory at HKUMed is a trailblazer in the realm of structural biology research, featuring cutting-edge instruments in Hong Kong. Since its soft launch in August 2023, the Laboratory has had over 60 users from more than 25 research groups, who are actively involved in over 40 projects covering a diverse range of topics including single particle analysis, tomography, protein-protein interactions, protein-DNA complexes, virus structures, bacteria, and small molecule studies.
Equipped with advanced microscopes such as the Krios 300kV TEM, Glacios 200kV TEM, Aquilos Cryo-FIB SEM, and Talos 120kV TEM, the Laboratory enables researchers to explore biological samples at near-atomic resolution and offers advanced sample freezing equipment for optimising sample quality for high-quality structural biology studies.
Beyond its instrumental capabilities, the Laboratory is committed to fostering a collaborative research environment by conducting training sessions, workshops, and seminars on Cryo-EM technology, highlighting the Laboratory’s pivotal role in driving scientific progress and shaping the future of structural biology.
Media enquiries
Please contact LKS Faculty of Medicine of The University of Hong Kong by email (medmedia@hku.hk).


Follow HKUMed